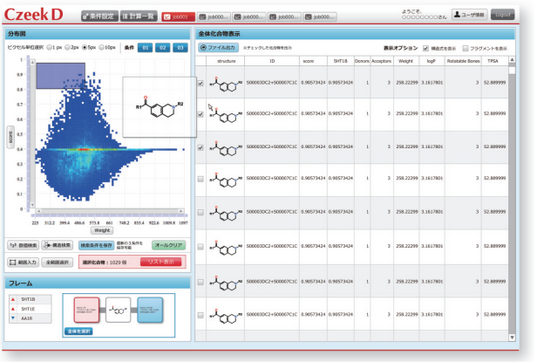
CzeekS
Optimization algorithm-inspired de-novo compound design system
Product Features
- Ease of use geared towards medicinal chemists
- Cloud computing service
- Efficient search of chemical space through the use of optimization algorithms
Optimization Step
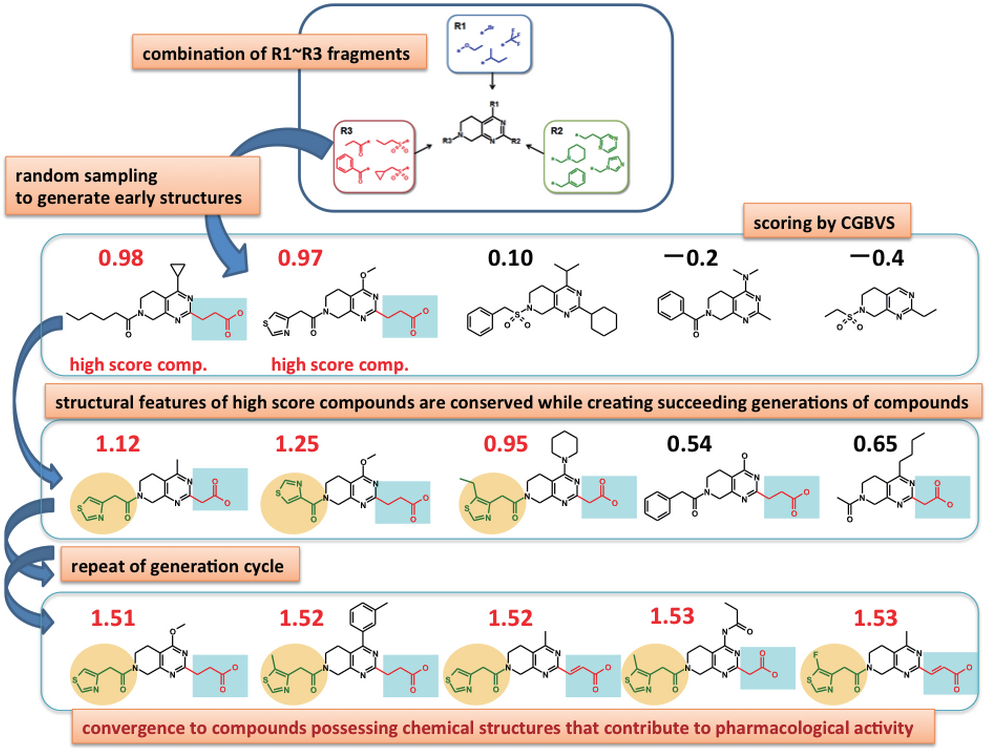 Fragment-based compound design
Fragment-based compound design
- Compounds are designed through the linkage of fragments based on the synthesis frame. A library of compounds that can be chemically synthesized is created using rules based on RECAP
RECAP – Retrosynthetic Combinatorial Analysis Procedure
- Frame setting
- Compounds are designed through the fitting of fragments to arbitrary frames.
- Construction of various chemical libraries after functional evaluation using CGBVS
- Multi-target screening via Chemical Genomics-Based Virtual Screening (CGBVS) enables the design of compounds with very high specificity, as well as, compounds with multiple protein targets.
Chemical Genomics-Based Virtual Screening
- CzeekS utilizes a machine learning method for virtual screening based on knowledge of chemical-genomics. High-speed virtual screening with high-accuracy is implemented in this system through a command line interface.
- Compound screening by multi-target prediction
- Scoring against multiple protein targets enables screening that takes the selectivity of compounds into consideration.
- Search for each compound’s target protein(s)
- Scoring of each compound against all the proteins included in the prediction model can be done facilitating the discovery of the target proteins.
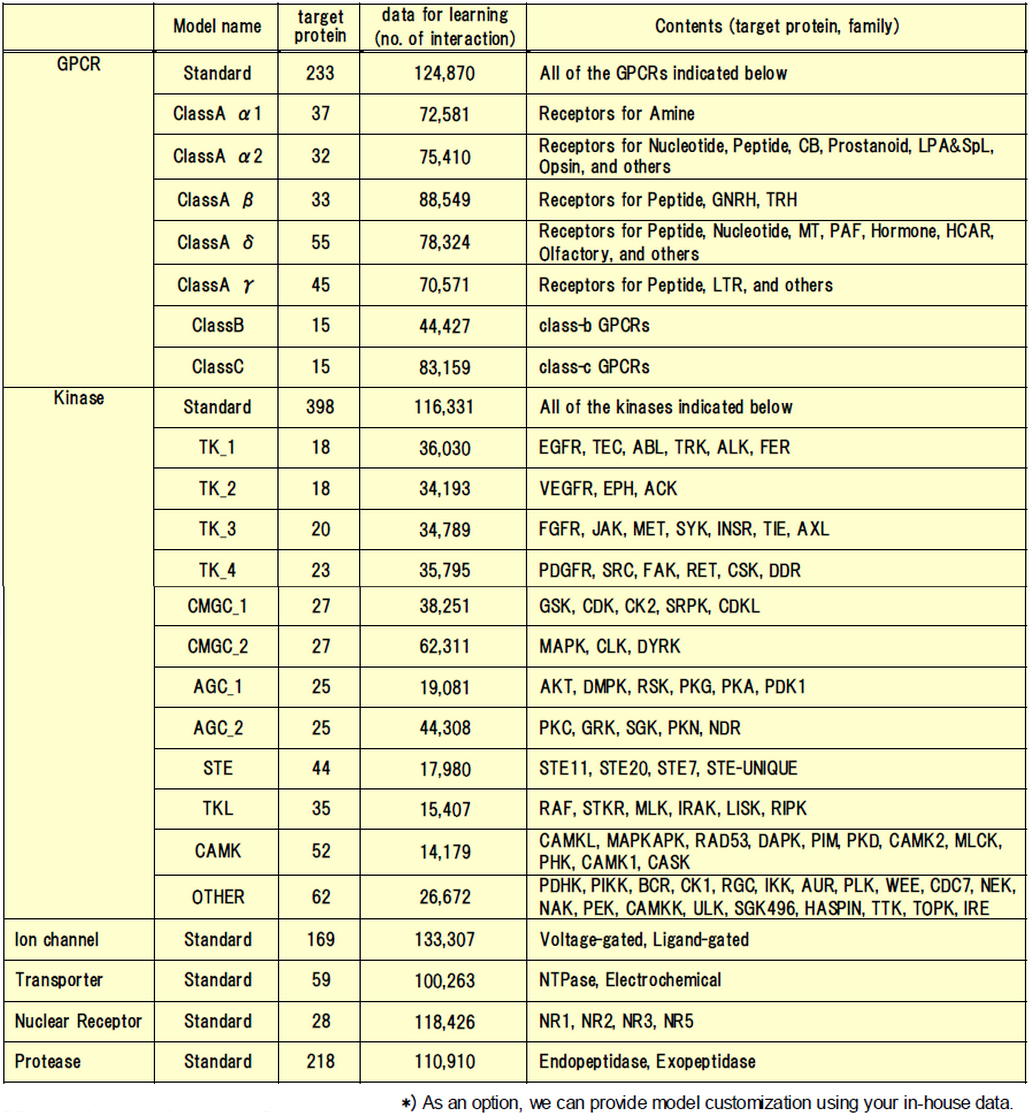
- A lineup of several prediction models
- In addition to standard prediction models (GPCR, Kinase, Ion channel, Transporter, Nuclear receptor, Protease), models focusing on subfamilies are also available. Clients can choose only the models that suit their purpose.
- In-house data can be used to create prediction models
- Clients’ data obtained from in vitro assays can be used to refine existing prediction models. The accuracy of the prediction model is expected to increase through the addition of new data.
- Compatibility with multicore processors (OpenMP)
- The ability of CzeekS to utilize multicore processors enables rapid CGBVS simulations.